Three CS Students Recognized By The Computing Research Association
For this year’s Outstanding Undergraduate Researcher Award, Payal Chandak, Sophia Kolak, and Yanda Chen were among students recognized by the Computing Research Association (CRA) for their work in an area of computing research.
 Payal Chandak
Payal Chandak
Finalist
Using Machine Learning to Identify Adverse Drug Effects Posing Increased Risk to Women
Payal Chandak Columbia University, Nicholas Tatonetti Columbia University
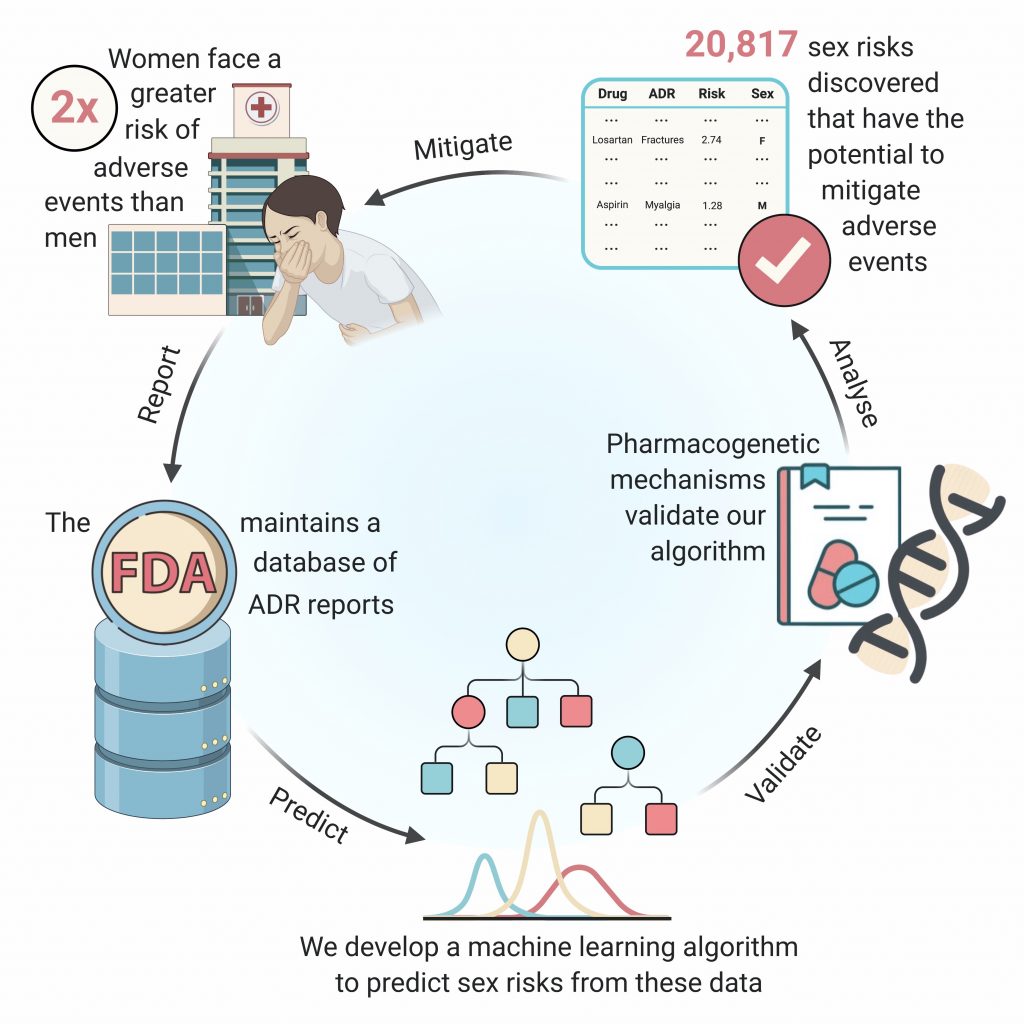
The researchers developed AwareDX – Analysing Women At Risk for Experiencing Drug toXicity – a machine learning algorithm that identifies and predicts differences in adverse drug effects between men and women by analyzing 50 years’ worth of reports in an FDA database. The algorithm automatically corrects for biases in these data that stem from an overrepresentation of male subjects in clinical research trials.
Though men and women can have different responses to medications – the sleep aid Ambien, for example, metabolizes more slowly in women, causing next-day grogginess – doctors may not know about these differences because most clinical trial data itself is biased toward men. This trickles down to impact prescribing guidelines, drug marketing, and ultimately, patients’ health. Unfortunately, pharmaceutical companies have a history of ignoring complex problems and clinical trials have singularly studied men, not even including women. As a result, there is a lot less information about how women respond to drugs compared to men. The research tries to bridge this information gap.
Buy Weed Seeds from premium breeders of the best marijuana strains. Discreetly ships across the USA with free weed seeds in every order.
 Sophia Kolak
Sophia Kolak
Finalist
It Takes a Village to Build a Robot: An Empirical Study of The ROS Ecosystem
Sophia Kolak Columbia University, Afsoon Afzal Carnegie Mellon University, Claire Le Goues Carnegie Mellon University, Michael Hilton Carnegie Mellon University, Christopher Steven Timperley Carnegie Mellon University
The Robot Operating System (ROS) is the most popular framework for robotics development. In this paper, the researchers conducted the first major empirical study of ROS, with the goal of understanding how developers collaborate across the many technical disciplines that coalesce in robotics.
Building a complete robot is a difficult task that involves bridging many technical disciplines. ROS aims to simplify development by providing reusable libraries, tools, and conventions for building a robot. Still, as building a robot requires domain expertise in software, mechanical, and electrical engineering, as well as artificial intelligence and robotics, ROS faces knowledge-based barriers to collaboration. The researchers wanted to understand how the necessity of domain-specific knowledge impacts the open-source collaboration model in ROS.
Virtually no one is an expert in every subdomain of robotics: experts who create computer vision packages likely need to rely on software designed by mechanical engineers to implement motor control. As a result, the researchers found that development in ROS is centered around a few unique subgroups each devoted to a different specialty in robotics (i.e. perception, motion). This is unlike other ecosystems, where competing implementations are the norm.
Detecting Performance Patterns with Deep Learning
Sophia Kolak Columbia University
Performance has a major impact on the overall quality of a software project. Performance bugs—bugs that substantially decrease run-time—have long been studied in software engineering, and yet they remain incredibly difficult for developers to handle. In this project, the researchers leveraged contemporary methods in machine learning to create graph embeddings of Python code that can be used to automatically predict performance.
Using un-optimized programming language concepts can lead to performance bugs and the researchers hypothesized that statistical language embeddings could help reveal these patterns. By transforming code samples into graphs that captured the control and data flow of a program, the researchers studied how various unsupervised embeddings of these graphs could be used to predict performance.
Implementing “sort” by hand as opposed to using the built-in Python sort function is an example of a choice that typically slows down a program’s run-time. When the researchers embedded the AST and data flow of a code snippet in Euclidean space (using DeepWalk), patterns like this were captured in the embedding and allowed classifiers to learn which structures are correlated with various levels of performance.
“I was surprised by how often research changes directions,” said Sophia Kolak. In both projects, they started out with one set of questions but answered completely different ones by the end. “It showed me that, in addition to persistence, research requires open-mindedness.”
 Yanda Chen
Yanda Chen
Honorable Mention
Cross-language Sentence Selection Via Data Augmentation and Rationale Training
Yanda Chen Columbia University, Chris Kedzie Columbia University, Suraj Nair University of Maryland, Petra Galuscakova University of Maryland, Rui Zhang Yale University, Douglas Oard University of Maryland, and Kathleen McKeown Columbia University
In this project, the researchers proposed a new approach to cross-language sentence selection, where they used models to predict sentence-level query relevance with English queries over sentences within document collections in low-resource languages such as Somali, Swahili, and Tagalog.
The system is used as part of cross-lingual information retrieval and query-focused summarization system. For example, if a user puts in a query word “business activity” and specifies Swahili as the language of source documents, then the system will automatically retrieve the Swahili documents that are related to “business activity” and produce short summaries that are then translated from Swahili to English.
A major challenge of the project was the lack of training data for low-resource languages. To tackle this problem, the researchers proposed to generate a relevance dataset of query-sentence pairs through data augmentation based on parallel corpora collected from the web. To mitigate the spurious correlations learned by the model, they proposed the idea of rationale training where they first trained a phrase-based statistical machine translation system and used the alignment information to provide additional supervision for the models.
The approach achieved state-of-the-art results on both text and speech across three languages – Somali, Swahili, and Tagalog.