Highlights
|
Variational Combinatorial Sequential Monte Carlo Methods for Bayesian Phylogenetic Inference
Antonio Khalil Moretti*,
Liyi Zhang*, Christian Naesseth, Hadiah Venner, David Blei,
Itsik Pe'er
Uncertainty in Artificial Intelligence, 2021
arxiv /
code
/
old slides /
talk /
bibtex
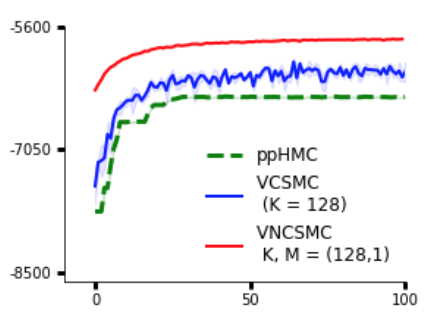
Bayesian phylogenetic inference is often conducted via local or sequential search over topologies and branch lengths using algorithms such as random-walk Markov chain Monte Carlo (MCMC) or Combinatorial Sequential Monte Carlo (CSMC). However, when MCMC is used for evolutionary parameter learning, convergence requires long runs with inefficient exploration of the state space. We introduce Variational Combinatorial Sequential Monte Carlo (VCSMC), a powerful framework that establishes variational sequential search to learn distributions over intricate combinatorial structures. We then develop nested CSMC, an efficient proposal distribution for CSMC and prove that nested CSMC is an exact approximation to the (intractable) locally optimal proposal. We use nested CSMC to define a second objective, VNCSMC which yields tighter lower bounds than VCSMC. We show that VCSMC and VNCSMC are computationally efficient and explore higher probability spaces than existing methods on a range of tasks.
|
|
Particle Smoothing Variational Objectives
Antonio Khalil Moretti*,
Zizhao Wang*,
Luhuan Wu*,
Iddo Drori,
Itsik Pe'er
European Conference on Artificial Intelligence, 2020
arxiv /
code
/
slides /
talk /
bibtex
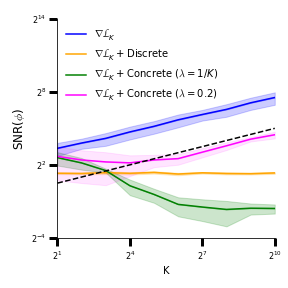
A body of recent work has focused on constructing a variational family of filtered distributions using Sequential Monte Carlo (SMC). Inspired by this work, we introduce Smoothing Variational Objectives (SVO), a novel backward simulation technique defined through a subsampling process. This augments the support of the proposal and boosts particle diversity. SMCs resampling step introduces challenges for standard VAE-style reparameterization due to the Categorical distribution. We prove that introducing bias by dropping this term from the gradient estimate or using Gumbel-Softmax mitigates the adverse effect on the signal-to-noise ratio. SVO consistently outperforms filtered objectives when given fewer Monte Carlo samples on three nonlinear systems of increasing complexity.
|
|
Nonlinear Evolution via Spatially-Dependent Linear Dynamics for Electrophysiology and Calcium Data
Daniel Hernandez Diaz,
Antonio Khalil Moretti,
Ziqiang Wei,
Shreya Saxena,
John Cunningham,
Liam Paninski
Neurons, Behavior, Data analysis, and Theory, 2020
arxiv
/
code
/
bibtex
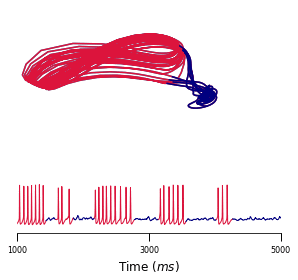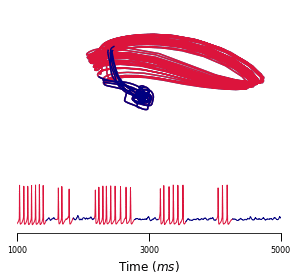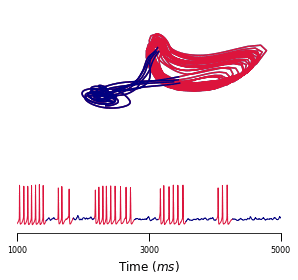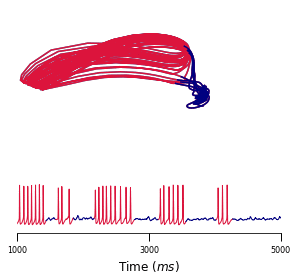
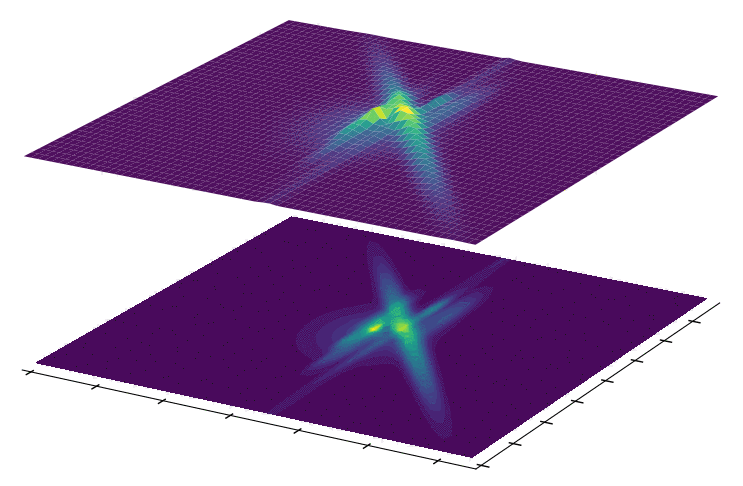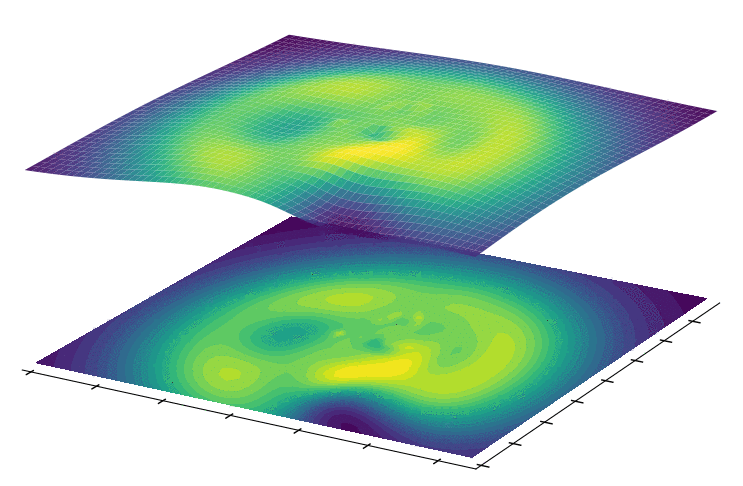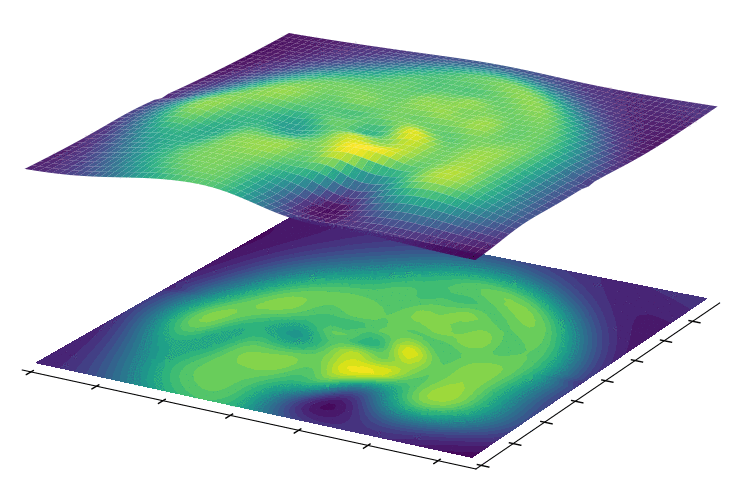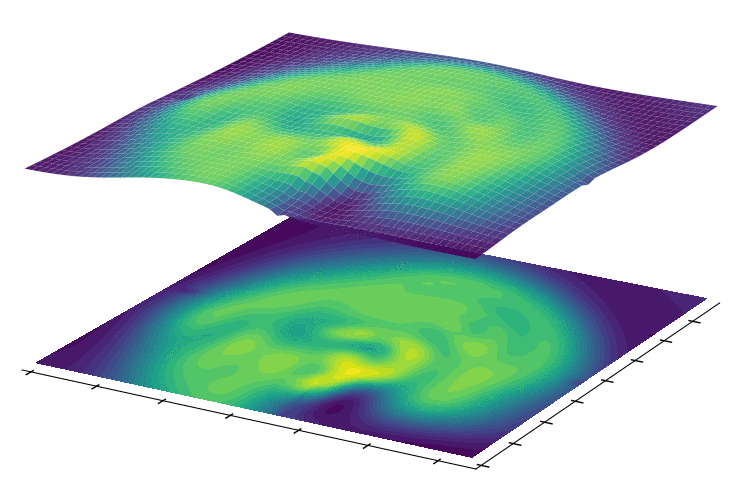
We propose a novel variational inference framework for the explicit modeling of time series, Variational Inference for Nonlinear Dynamics (VIND), that is able to uncover nonlinear observation and transition functions from sequential data. The framework includes a structured approximate posterior, and an algorithm that relies on the fixed-point iteration method to find the best estimate for latent trajectories. We apply VIND to electrophysiology, single-cell voltage and widefield optical imaging datasets with state-of-the-art results in reconstruction error. In single-cell voltage data, VIND finds a 5D latent space, with variables akin to those of Hodgkin-Huxley-like models. The quality of learned dynamics is further quantified by using it to predict future neural activity. VIND excels in this task, in some cases substantially outperforming state-of-the-art methods.
|
|
Autoencoding Topographic Factors
Antonio Khalil Moretti*,
Andrew Atkinson Stirn*,
Gabriel Marks,
Itsik Pe'er
Journal of Computational Biology, 2019
arxiv
/
code
/
bibtex
Topographic factor models separate a set of overlapping singals into spatially localized source functions without knowledge of the original signals or the mixing process. We propose Auto-Encoding Topographic Factors (AETF), a novel variational inference scheme that does not require sources to be held constant across locations on the lattice. Model parameters scale independently of dataset size making it possible to perform inference on temporal sequences of large 3D image matrices. AETF is evaluated on both simulations and on deep generative models of functional magnetic resonance imaging data and is shown to outperform existing Topographic factor models in reconstruction error.
|
|
Publications & Preprints
-
Variational Combinatorial Sequential Monte Carlo Methods for Bayesian Phylogenetic Inference
Moretti, A.*, Zhang, L.*, Naesseth, C., Venner, H., Blei, D., Pe'er, I.
Uncertainty in Artificial Intelligence, 2021
-
Variational Combinatorial Sequential Monte Carlo for Bayesian Phylogenetic Inference
Moretti, A., Zhang, L., Pe'er, I.
Machine Learning in Computational Biology, 2020
Selected for Oral Presentation (15% Acceptance Rate)
-
Variational Objectives for Markovian Dynamics with Backward Simulation
Moretti, A.*, Wang, Z.*, Wu, L.*, Drori, I., Pe'er, I.
European Conference on Artificial Intelligence, 2020
-
Nonlinear Evolution via Spatially-Dependent Linear Dynamics for Electrophysiology and Calcium Data
Hernandez, D., Moretti, A., Wei, Z., Saxena, S., Cunningham, J., Paninski, L.
Neurons, Behavior, Data analysis and Theory, 2020
-
Accurate Protein Structure Prediction by Embeddings and Deep Learning Representations
Drori, I., Thaker, D., Srivatsa, A., Jeong, D., Wang, Y., Nan, L., Wu, F., Leggas, D., Lei, J., Lu, W., Fu, W., Gao, Y., Karri, S., Kannan, A., Moretti, A., Keasar, C., Pe'er, I.
Machine Learning in Computational Biology, 2019
-
Particle Smoothing Variational Objectives
Moretti, A.*, Wang, Z.*, Wu, L.*, Drori, I., Pe'er, I.
arXiv preprint arXiv:1909.09734
-
Auto-Encoding Topographic Factors
Moretti, A.*, Stirn, A.*, Marks, G., Pe'er, I.
Journal of Computational Biology, 2019
-
The Impact of Comprehensive Case Management on HIV Client Outcomes
Brennan-Ing M., Seidel L., Rodgers L., Ernst J., Wirth D., Tietz D., Moretti, A., Karpiak, S.
Plos One, 2016
-
AutoML using Metadata Language Embeddings
Drori, I., Liu, L., Nian, Y., Koorathota, S., Li, J., Moretti, A., Freire, J., Udell, M.
NeurIPS Workshops, 2019
-
Smoothing Nonlinear Variational Objectives with Sequential Monte Carlo
Moretti, A.*, Wang, Z.*, Wu, L., Pe'er, I.
ICLR Workshops, 2019
-
Nonlinear Variational Inference for Neural Data
Hernandez, D., Moretti, A., Wei, Z., Saxena, S., Cunningham, J., Paninski, L.
Computational and Systems Neuroscience, 2019
-
Auto-Encoding Topographic Factors
Moretti, A.*, Stirn, A.*, Pe'er, I.
ICML Workshops, 2018
Selected for Oral Presentation (17% Acceptance Rate) and Best Poster Award
-
Mining Student Ratings and Course Contents for CS Curriculum Decisions
Moretti, A., Gonzalez-Brenes, J., McKnight, K. and Salleb-Aouissi, A.
KDD Workshops, 2014Spotlight Talk
|