 |
Pablo A. Duboue - Kathleen R. McKeown
Computer Science Department
Columbia University
10027, New York, NY, USA
{pablo,kathy}@cs.columbia.edu
In a language generation system, a content planner typically uses one or more ``plans'' to represent the content to be included in the output and the ordering between content elements. Some researchers rely on generic planners (e.g., [6]) for this task, while others use plans based on Rhetorical Structure Theory (RST) (e.g., [3,17,9]) or schemas (e.g., [14,13]). In all cases, constraints on application of rules (e.g., plan operators), which determine content and order, are usually hand-crafted, sometimes through manual analysis of target text.
In this paper, we present a method for learning the basic patterns contained within a plan and the ordering among them. As training data, we use semantically tagged transcripts of domain experts performing the task our system is designed to mimic, an oral briefing of patient status after undergoing coronary bypass surgery. Given that our target output is spoken language, there is some level of variability between individual transcripts. It is difficult for a human to see patterns in the data and thus supervised learning based on hand-tagged training sets can not be applied. We need a learning algorithm that can discover ordering patterns in apparently unordered input.
We based our unsupervised learning algorithm on techniques used in computational genomics [7], where from large amounts of seemingly unorganized genetic sequences, patterns representing meaningful biological features are discovered. In our application, a transcript is the equivalent of a sequence and we are searching for patterns that occur repeatedly across multiple sequences. We can think of these patterns as the basic elements of a plan, representing small clusters of semantic units that are similar in size, for example, to the nucleus-satellite pairs of RST.1 By learning ordering constraints over these elements, we produce a plan that can be expressed as a constraint-satisfaction problem. In this paper, we focus on learning the plan elements and the ordering constraints between them. Our system uses combinatorial pattern matching [19] combined with clustering to learn plan elements. Subsequently, it applies counting procedures to learn ordering constraints among these elements.
Our system produced a set of 24 schemata units, that we call ``plan elements''2, and 29 ordering constraints between these basic plan elements, which we compared to the elements contained in the orginal hand-crafted plan that was constructed based on hand-analysis of transcripts, input from domain experts, and experimental evaluation of the system [15].
The remainder of this article is organized as follows: first the data used in our experiments is presented and its overall structure and acquisition methodology are analyzed. In Section 3 our techniques are described, together with their grounding in computational genomics. The quantitative and qualitative evaluation are discussed in Section 4. Related work is presented in Section 5. Conclusions and future work are discussed in Section 6.
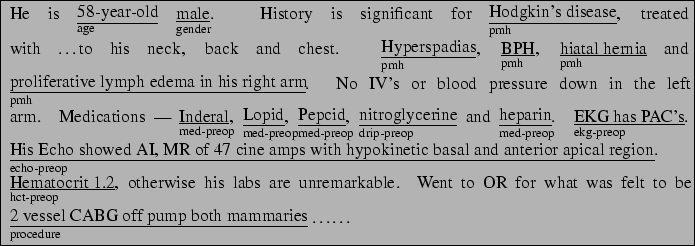
Our research is part of MAGIC [5,15], a system that is designed to produce a briefing of patient status after undergoing a coronary bypass operation. Currently, when a patient is brought to the intensive care unit (ICU) after surgery, one of the residents who was present in the operating room gives a briefing to the ICU nurses and residents. Several of these briefings were collected and annotated for the aforementioned evaluation. The resident was equipped with a wearable tape recorder to tape the briefings, which were transcribed to provide the base of our empirical data. The text was subsequently annotated with semantic tags as shown in Figure 1. The figure shows that each sentence is split into several semantically tagged chunks. The tag-set was developed with the assistance of a domain expert in order to capture the different information types that are important for communication and the tagging process was done by two non-experts, after measuring acceptable agreement levels with the domain expert (see [15]). The tag-set totalled over 200 tags. These 200 tags were then mapped to 29 categories, which was also done by a domain expert. These categories are the ones used for our current research.
From these transcripts, we derive the sequences of semantic tags for
each transcript. These sequences constitute the input and working
material of our analysis, they are an average length of 33 tags per
transcript (![]() ,
, ![]() ,
, ![]() ). A tag-set
distribution analysis showed that some of the categories dominate the
tag counts. Furthermore, some tags occur fairly regularly towards either the
beginning (e.g., date-of-birth) or the end (e.g.,
urine-output) of the transcript, while others (e.g.,
intraop-problems) are spread more or less evenly
throughout.
). A tag-set
distribution analysis showed that some of the categories dominate the
tag counts. Furthermore, some tags occur fairly regularly towards either the
beginning (e.g., date-of-birth) or the end (e.g.,
urine-output) of the transcript, while others (e.g.,
intraop-problems) are spread more or less evenly
throughout.
Getting these transcripts is a highly expensive task involving the cooperation and time of nurses and physicians in the busy ICU. Our corpus contains a total number of 24 transcripts. Therefore, it is important that we develop techniques that can detect patterns without requiring large amounts of data.
During the preliminary analysis for this research, we looked for techniques to deal with analysis of regularities in sequences of finite items (semantic tags, in this case). We were interested in developing techniques that could scale as well as work with small amounts of highly varied sequences.
Computational biology is another branch of computer science that has this problem as one topic of study. We focused on motif detection techniques as a way to reduce the complexity of the overall setting of the problem. In biological terms, a motif is a small subsequence, highly conserved through evolution. From the computer science standpoint, a motif is a fixed-order pattern, simply because it is a subsequence. The problem of detecting such motifs in large databases has attracted considerable interest in the last decade (see [10] for a recent survey). Combinatorial pattern discovery, one technique developed for this problem, promised to be a good fit for our task because it can be parameterized to operate successfully without large amounts of data and it will be able to identify domain swapped motifs: for example, given a-b-c in one sequence and c-b-a in another. This difference is central to our current research, given that order constraints are our main focus. TEIRESIAS [19] and SPLASH [4] are good representatives of this kind of algorithm. We used an adaptation of TEIRESIAS.
The algorithm can be sketched as follows: we apply combinatorial pattern discovery (see Section 3.1) to the semantic sequences. The obtained patterns are refined through clustering (Section 3.2). Counting procedures are then used to estimate order constraints between those clusters (Section 3.3).
Using the previous definitions, the algorithm reduces to the problem
of, given a set of sequences, ![]() ,
, ![]() , a minimum windowsize, and a
support threshold, finding maximal
, a minimum windowsize, and a
support threshold, finding maximal
![]() -patterns with at least a support of support
threshold. Our implementation can be sketched as follows:
-patterns with at least a support of support
threshold. Our implementation can be sketched as follows:
After the detection of patterns is finished, the number of patterns is relatively large. Moreover, as they have fixed length, they tend to be pretty similar. In fact, many tend to have their support from the same subsequences in the corpus. We are interested in syntactic similarity as well as similarity in context.
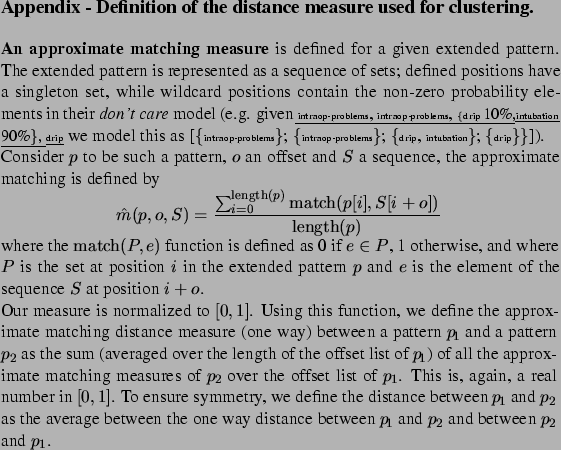
A convenient solution was to further cluster the patterns, according to an approximate matching distance measure between patterns, defined in an appendix at the end of the paper.
We use agglomerative clustering with the distance between clusters defined as the maximum pairwise distance between elements of the two clusters. Clustering stops when no inter-cluster distance falls below a user-defined threshold.
Each of the resulting clusters has a single pattern represented by the centroid of the cluster. This concept is useful for visualization of the cluster in qualitative evaluation.
The last step of our algorithm measures the frequencies of all possible order constraints among pairs of clusters, retaining those that occur often enough to be considered important, according to some relevancy measure. We also discard any constraint that it is violated in any training sequence. We do this in order to obtain clear-cut constraints. Using the number of times a given constraint is violated as a quality measure is a straight-forward extension of our framework. The algorithm proceeds as follows: we build a table of counts that is updated every time a pair of patterns belonging to particular clusters are matched. To obtain clear-cut constraints, we do not count overlapping occurrences of patterns.
From the table of counts we need some relevancy measure, as the
distribution of the tags is skewed. We use a simple heuristic to
estimate a relevancy measure over the constraints that are never
contradicted. We are trying to obtain an estimate of
The obtained estimates, ![]() and
and ![]() , will in general
yield different numbers. We use the arithmetic mean between both,
, will in general
yield different numbers. We use the arithmetic mean between both,
![]() , as the final estimate for each constraint. It
turns out to be a good estimate, that predicts accuracy of the
generated constraints (see Section 4).
, as the final estimate for each constraint. It
turns out to be a good estimate, that predicts accuracy of the
generated constraints (see Section 4).
We use cross validation to quantitatively evaluate our results and a comparison against the plan of our existing system for qualitative evaluation.
We evaluated two items: how effective the patterns and constraints learned were in an unseen test set and how accurate the predicted constraints were. More precisely:
Using 3-fold cross-validation for computing these metrics, we obtained
the results shown in Table 1 (averaged over 100
executions of the experiment).
The different parameter settings were defined as follows: for the
motif detection algorithm
![]() and
support threshold of 3. The algorithm will normally find
around 100 maximal motifs. The clustering algorithm used a relative
distance threshold of 3.5 that translates to an actual treshold of 120
for an average inter-cluster distance of 174. The number of produced
clusters was in the order of the 25 clusters or so. Finally, a
threshold in relevancy of 0.1 was used in the constraint learning
procedure. Given the amount of data available for these experiments
all these parameters were hand-tunned.
and
support threshold of 3. The algorithm will normally find
around 100 maximal motifs. The clustering algorithm used a relative
distance threshold of 3.5 that translates to an actual treshold of 120
for an average inter-cluster distance of 174. The number of produced
clusters was in the order of the 25 clusters or so. Finally, a
threshold in relevancy of 0.1 was used in the constraint learning
procedure. Given the amount of data available for these experiments
all these parameters were hand-tunned.
The system was executed using all the available information, with the same parametric settings used in the quantitative evaluation, yielding a set of 29 constraints, out of 23 generated clusters.
These constraints were analyzed by hand and compared to the existing content-planner. We found that most rules that were learned were validated by our existing plan. Moreover, we gained placement constraints for two pieces of semantic information that are currently not represented in the system's plan. In addition, we found minor order variation in relative placement of two different pairs of semantic tags. This leads us to believe that the fixed order on these particular tags can be relaxed to attain greater degrees of variability in the generated plans. The process of creation of the existing content-planner was thorough, informed by multiple domain experts over a three year period. The fact that the obtained constraints mostly occur in the existing plan is very encouraging.
 |
As explained in [10], motif detection is usually targeted with alignment techniques (as in [7]) or with combinatorial pattern discovery techniques such as the ones we used here. Combinatorial pattern discovery is more appropriate for our task because it allows for matching across patterns with permutations, for representation of wild cards and for use on smaller data sets.
Similar techniques are used in NLP. Alignments are widely used in MT, for example [16], but the crossing problem is a phenomenon that occurs repeatedly and at many levels in our task and thus, this is not a suitable approach for us.
Pattern discovery techniques are often used for information extraction (e.g., [20,8]), but most work uses data that contains patterns labelled with the semantic slot the pattern fills. Given the difficulty for humans in finding patterns systematically in our data, we needed unsupervised techniques such as those developed in computational genomics.
Other stochastic approaches to NLG normally focus on the problem of sentence generation, including syntactic and lexical realization (e.g., [12,1,11]). Concurrent work analyzing constraints on ordering of sentences in summarization found that a coherence constraint that ensures that blocks of sentences on the same topic tend to occur together [2]. This results in a bottom-up approach for ordering that opportunistically groups sentences together based on content features. In contrast, our work attempts to automatically learn plans for generation based on semantic types of the input clause, resulting in a top-down planner for selecting and ordering content.
In this paper we presented a technique for extracting order constraints among plan elements that performs satisfactorily without the need of large corpora. Using a conservative set of parameters, we were able to reconstruct a good portion of a carefully hand-crafted planner. Moreover, as discussed in the evaluation, there are several pieces of information in the transcripts which are not present in the current system. From our learned results, we have inferred placement constraints of the new information in relation to the previous plan elements without further interviews with experts.
Furthermore, it seems we have captured order-sensitive information in the patterns and free-order information is kept in the don't care model. The patterns, and ordering constraints among them, provide a backbone of relatively fixed structure, while don't cares are interspersed among them. This model, being probabilistic in nature, means a great deal of variation, but our generated plans should have variability in the right positions. This is similar to findings of floating positioning of information, together with oportunistic rendering of the data as used in STREAK [21].
We are planning to use these techniques to revise our current content-planner and incorporate information that is learned from the transcripts to increase the possible variation in system output.
The final step in producing a full-fledged content-planner is to add semantic constraints on the selection of possible orderings. This can be generated through clustering of semantic input to the generator.
We also are interested in further evaluating the technique in an unrestricted domain such as the Wall Street Journal (WSJ) with shallow semantics such as the WordNet top-category for each NP-head. This kind of experiment may show strengths and limitations of the algorithm in large corpora.