An interview with John Paparrizos: Hunting early signs of pancreatic cancer in web searches
With few warning signs and no screening test, pancreatic cancer is usually not diagnosed until it is well advanced; 75% of pancreatic cancer patients die within a year of a diagnosis. The prognosis improves if the disease is caught early, but the cancer’s nonspecific symptoms (bloating, abdominal or back pain) are not initially concerning. Doctors will want to rule out more likely diseases first before ordering expensive lab tests; adding to the difficulty, doctors often lack a full medical history for patients they see only intermittently or even for the first time and patients may have visited other doctors or facilities. Doctors in other words have very little data to go on.
Search engines however collect a great deal of data, and as more people turn to the web for health-related information, they are constructing a deep and broad data set of medical histories that may contain clues as to how diseases emerge.
In a study started last summer, researchers Eric Horvitz and Ryen White of Microsoft, both recognized experts in the field of information retrieval, along with Columbia PhD student John Paparizzos looked through 18 months of US Bing search data to see if they could detect patterns of symptoms in pancreatic cancer before people were diagnosed with the disease. In 5–15% of such cases, they could, and with extremely few false positives (as low as 1 in 100,000).
A paper detailing their results was the August 2016 cover article for The Journal of Oncology Practice and widely covered in the popular press. In this interview, John Paparizzos discusses the methodology behind those results and more recent work presented at KDD in August.

CS PhD student
(advisor Luis Gravano)
How did you know what users had been diagnosed with pancreatic cancer?
We were very careful. We looked for people whose first-person searches indicated they had the disease: “I was just diagnosed with pancreatic cancer,” “why did I get cancer in pancreas,” and “I was told I have pancreatic cancer what to expect.” We then worked backward in time through previous searches made by these people to see if they had previously searched for symptoms associated with the disease—bloating or abdominal discomfort, unexplained weight loss, yellowing skin. From 9.2 million users with searches relevant to pancreatic cancer disease and symptoms, we focused on 3203 cases matching the diagnostic pattern.
It’s important to point out that this data was completely anonymized. Users were identified through a unique ID; we had no names or personal information about them. Just search queries.
You study computer science. How did you come to work on a medical problem?
Before starting an internship last summer at Microsoft, I talked by phone with Eric and Ryen on possible projects. They have spent several years studying how people search online, particularly how people use the web for health purposes. The idea this time was to do some type of prediction through analysis of patients’ query logs over time for a specific disease. Pancreatic cancer was a good candidate; it’s a fast-developing cancer with few overt symptoms. Search logs could conceivably provide early warning of the disease, something that doesn’t exist now.
My particular focus was to develop an approach to capture characteristics over time in the user behavior—as expressed in search query logs—that would discriminate users as early as possible to those who might experience pancreatic cancer and those who simply explore pancreatic cancer.
Since human web searches contain a lot of noise, we spent a great deal of time cleaning and annotating data before inputting it into classifiers that separate out the true signals. We excluded searches where people were looking up symptoms for general knowledge or on the behalf of someone else. We excluded idiomatic phrases that might get mistaken for real symptoms, like “I’m sick of hearing about the Kardashians.” Particular to this problem were queries generated by Steve Jobs’s death from pancreatic cancer.
Was this the first time you had worked with medical data?
Yes. It was new to me and it required learning about the disease of pancreatic cancer and its symptoms, which were all described differently also. Medical records are usually clean, formatted in templates, and reflect the precise vocabulary of doctors working in the field, but people will describe those symptoms in very different terms, even using slang. We had to do a two-level ontology of terms for symptoms and their synonyms.
The distribution of the data was different. Normally you are building classifiers where the positive and the negative cases are in balance. With this project, maybe 1 in 10,000 people will have this disease. A high misprediction rate could unnecessarily alert millions of users.
There were issues of scale. In an academic environment we might be running analysis involving 100 or 1000 users, not millions. When you’re dealing with at least 3 orders of magnitude more, there is a lot of engineering involved along with the research task.
You knew from people’s previous searches who had cancer. Could you take this model and apply to another data set where it’s not known whether people have the cancer or not?
Our KDD paper presented last month describes a similar experiment. Where our first paper describes training a model on users between October 2013 to May 2015, our KDD paper describes how we took this model and applied it on a new set of users from August 2014 to January 2016—users not used in training. We wanted to see if we could predict which searchers will later input first-person queries for the disease, which we could do at about the same accuracy as before.
The second paper also extended the methodology. In addition to looking for people with first person searches—“I have this cancer”—the second paper goes an extra step and looks for users who searched for the specific treatments for this disease—Whipple procedure, pancreaticoduodenectomy, neoadjuvant therapy—that only someone with first-hand knowledge of the disease would know.
We got slightly better results because the data was cleaner, and this raised our accuracy level from 90% to 92%.
What is the possibility that web searches will actually be used to screen for cancer?
Our goal was to demonstrate the feasibility of mining medical histories of a large population of people to successfully see symptoms at an early stage. We did this using anonymized search data, but it’s not practical in the real world. We don’t have names so we can’t contact individuals—which would raise massive privacy issues in any case.
What may be feasible is to link our method to a hospital or clinical group where patients allow their data to be shared and combined. For this, we need help from doctors working directly with patients, and it’s why we published first in a medical journal; we wanted to engage the medical community with the prospect of using nontraditional web searches in conjunction with standard practices as a way to do low-cost, passive surveillance to flag hidden problems and improve cancer screenings.
Our system won’t do the diagnosis—that is for the doctor to do—but it pulls together more clues and gives more warning that something serious might be developing, and to perhaps recommend that a specific patient gets a certain test or suggest a meeting with a physician.
We want doctors to see what’s possible when you have more data; you learn more about a single patient by having that patient’s data centralized in one spot and seeing what symptoms they had months ago, but you also learn more about the disease. For example, we found in examining the histories of many pancreatic cancers that symptoms appearing in particular order—for example, indigestion before abdominal pain—correlate positively with the experiential user cases.
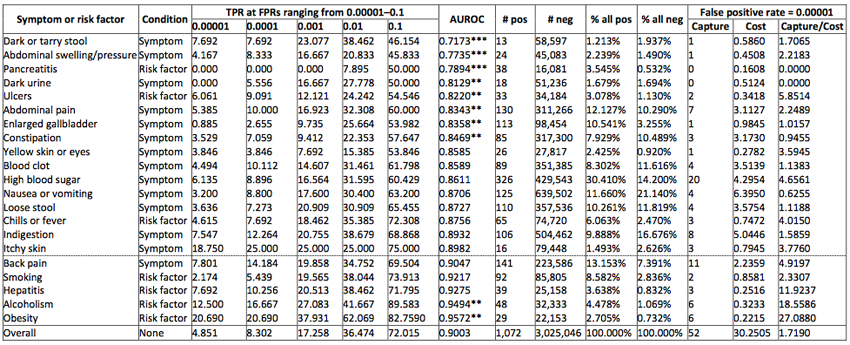
To better understand when such web-based health surveillance approach could be applied and what is the impact of the presence of particular risk factors or symptoms on the performance of the model, we have performed an analysis of the performance of our model when conditioned on users with specific symptoms or risk factors. We found that if we focus our predictions on users who search for risk factors such as alcoholism or obesity many more users would benefit from a prediction than would be mistakenly alerted.
Will you continue to work with medical data sets? Did this intrigue you a lot to work in this field?
Perhaps. It’s an extremely interesting application. In our field, you don’t get to see a direct application to users. We build a new model or classier or algorithm, but often it’s not linked to such a concrete application. This is an excellent example of how we can use science and what we are learning in computer science for social good.
Posted: 9/13/2016

